This material is part of a series of blog posts about using Julia from Python (Soon). The idea was initially presented internally at the Netherlands eScience Center(See the slides).
Post links:
- https://blog.esciencecenter.nl/how-to-call-julia-code-from-python-8589a56a98f2
- https://blog.esciencecenter.nl/speed-up-your-python-code-using-julia-f97a6c155630
- Soon
- We read Patrick's blog post about improving the reading of irregular files.
- Patrick has a Python (Pandas) code that is slow.
- Using some packages, he moves the reading and parsing to C++.
- We decided to try to replace C++ with Julia to check:
- How easy/hard it is
- How much improvement can be gained with a basic Julia code;
- How much further improvement can be gained with an optimized Julia code.
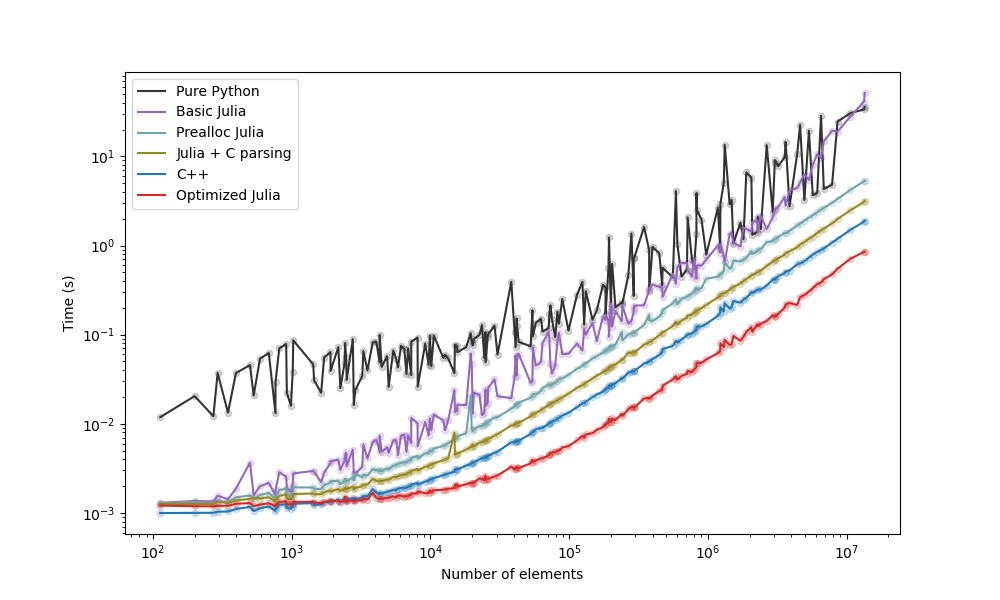
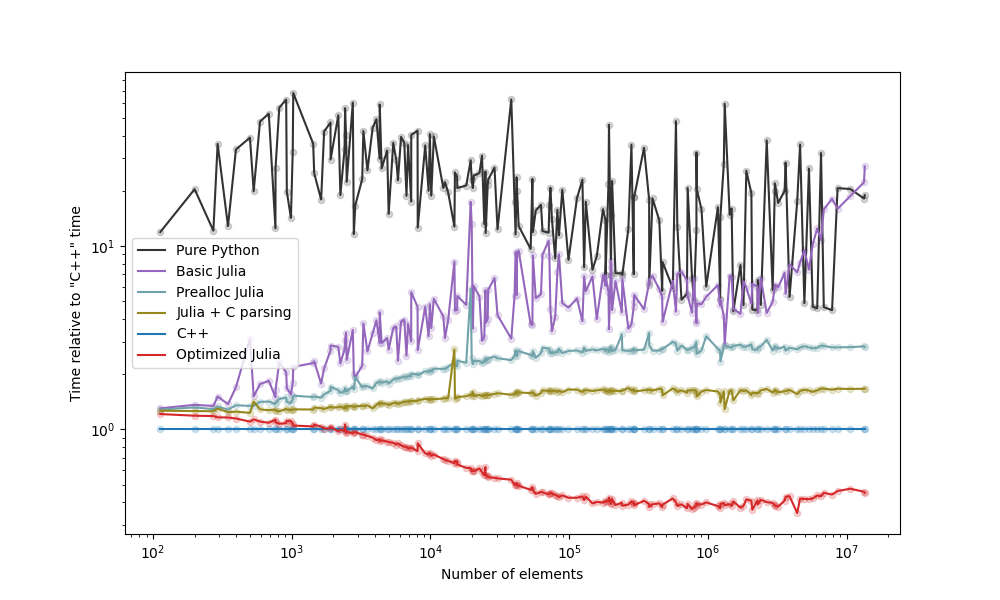
The strategies we examined are below, with a plot with the comparison following it:
- Python with Pandas, as seen in Patrick's post. label: "Pure Python".
- Python with reading and parsing in C++, as seen in Patrick's post. label: "C++".
- Python with reading and parsing in Julia, in 4 different versions:
- Basic Julia version with mostly disregard for efficiency, label="Basic Julia".
- Julia version trying to improve memory usage. label: "Prealloc Julia".
- Julia version where the elements are read with
fscanffrom C. label: "Julia + C parsing". - Julia version reading the file as bytes and manually walking through the bytes. label: "Optimized Julia".
Take-aways (see blog post):
- The "Prealloc Julia" strategy is already an improvement over the "Pure Python" strategy.
- The "Optimized Julia" strategy is faster than the "C++" strategy.
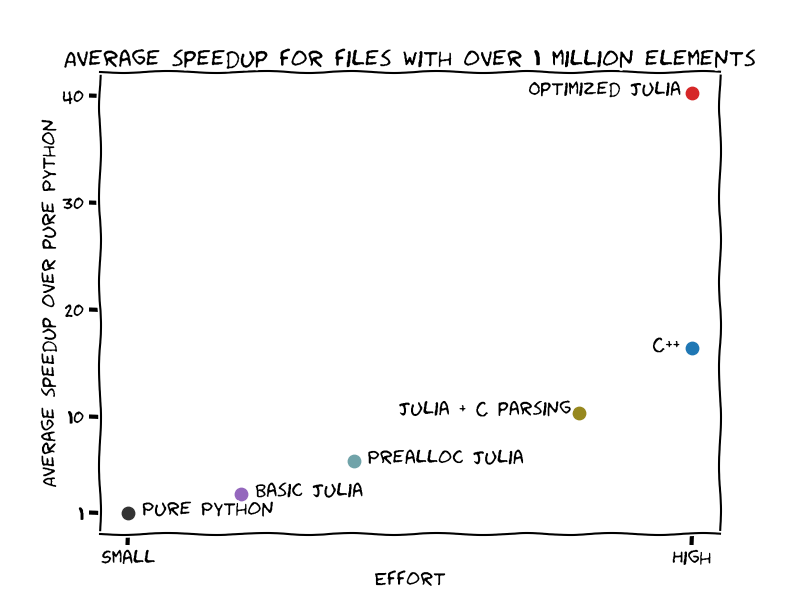
- If you don't know Julia nor C++, moving the slow code to Julia yields benefits faster and with less effort.
The image below shows the speedup gain over the effort to get there:
docker build --tag jl-from-py:<VERSION>-
Download dataset and store in a folder called
dataset. -
Get the image with
docker pull abelsiqueira/faster-python-with-julia-blogpost:post3
-
Run it with
docker run --rm --volume "$PWD/dataset:/app/dataset" --volume "$PWD/out:/app/out" abelsiqueira/faster-python-with-julia-blogpost:post3
-
You will find the outputs in the
out/folder.
The execution of this script with default options took about 45 minutes on a Dell Precision 5530 with the Intel chip i7-8850H (2.6GHz) and 16GiB of RAM.
The docker runs the script src/main.py that runs run_experiments.py and run_analysis.py.
--folder FOLDER: Set the dataset folder. (Default:dataset).--max-num-files N: Maximum number of files to read from can be used to limit the experiment. The files are traversed in sorted name order. Use 0 or a negative number to run all. (Default:0).--skip-after X: Time threshold in seconds to skip the tests of a specific version. If the threshold is reached twice, that version is skipped in the additional tests. (Default:0).--skip VALUE1 [VALUE2 ...]: List of versions to skip. Valid values:python,cpp,julia_basic,julia_c,julia_prealloc,julia_opt.