This is a pipeline for Bristol-Myers Squibb – Molecular Translation by Vadim Timakin and Maksim Zhdanov. We got bronze medals in this competition. Significant part of code was originated from Y.Nakama's notebook
This competition was about image-to-text translation of images with molecular skeletal strucutures to InChI chemical formula identifiers. Example of a text identifier:

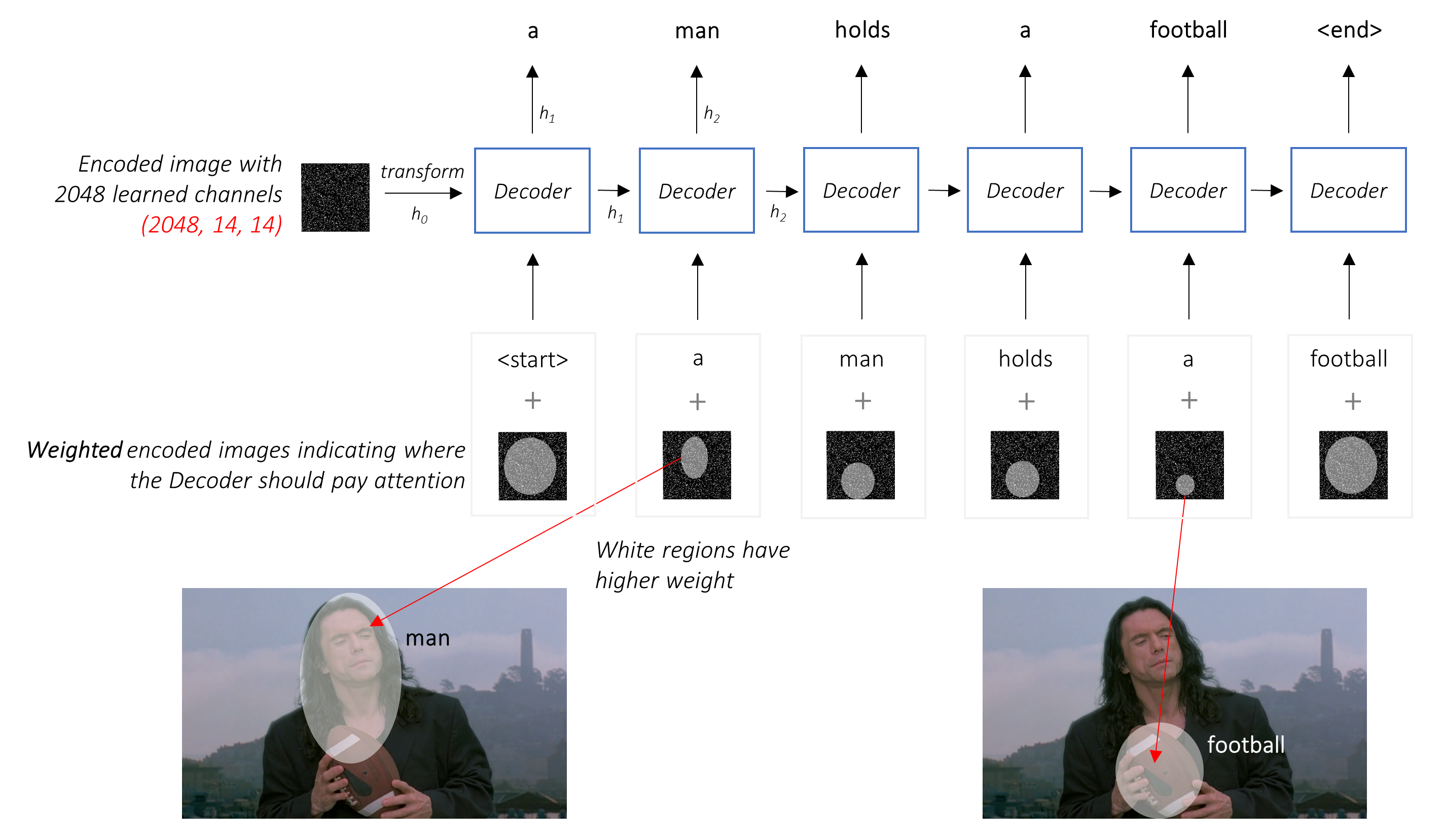
Most participants used CNN encoder to acquire features with decoder (LSTM/GRU/Transformer) to get text sequences.
That's a casual approach to image captioning problem.
RDKit is an open source toolkit for cheminformatics and it was quite useful while solving the problem. When we trained our first model, it scored around 7-8 on public leaderboard and we decided to make pseudo-labelling on test data. However, in common scenario you get a significant amount of wrong predictions in your extended training set from pseudo-labelling. With RDKit we validated all of our predicted formulas and select around 800k correct samples. Lack of wrong labels in pseudo labels improved the score.
This notebook tells about InChI normalization
Finally, we blended ~20 predictions from 2 models (mostly from different epochs) using RDKit validation to choose only formulas which have possible InChI structure.