Hi All,
Has anyone come across the issue described below? I'd appreciate any direction to help resolve this.
System information
OS Platform and Distribution: Windows 11 Home
Sklearn-genetic version: 0.5.1
deap version: 1.3.3
Scikit-learn version: 1.1.2
Python version: 3.8.13
Describe the bug
When running my pipeline to tune hyperparameters, this error occurs intermittently.
AssertionError: Assigned values have not the same length than fitness weights
I'm running TuneSearchCV (package tune-sklearn) to tune various hyperparameters of my pipeline below in this example, but have also encountered the error frequently when using GASearchCV (package sklearn-genetic-opt, also based on deap) :
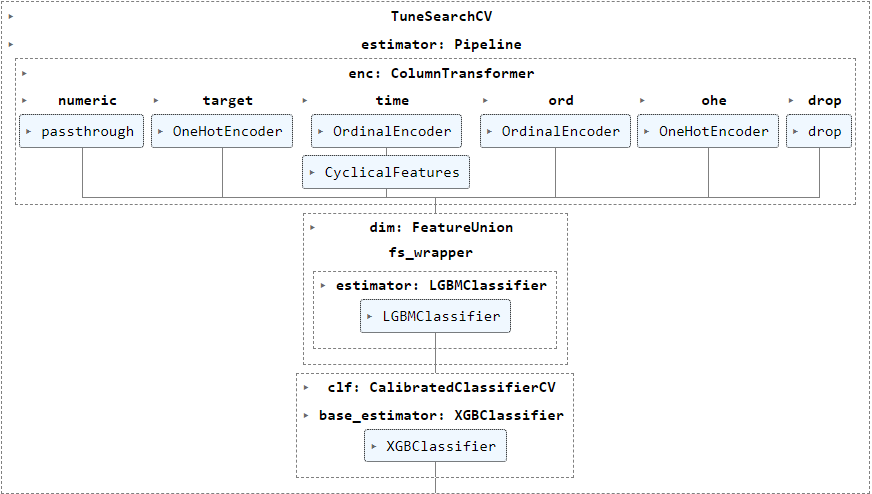
The following param_grid (generated using BayesSearchCV to show real information instead of the objects) show categorical values for various transformers steps enc__numeric, enc__target, enc__time__cyclicity and dim__fs_wrapper besides numerical parameter ranges for clf__base_estimator.
{'enc__numeric': Categorical(categories=('passthrough',
SmartCorrelatedSelection(selection_method='variance', threshold=0.9),
SmartCorrelatedSelection(cv='skf5',
estimator=LGBMClassifier(learning_rate=1.0,
max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1),
selection_method='model_performance', threshold=0.9)), prior=None),
'enc__target': Categorical(categories=(MeanEncoder(ignore_format=True), TargetEncoder(), MEstimateEncoder(),
WoEEncoder(ignore_format=True), PRatioEncoder(ignore_format=True),
BayesianTargetEncoder(columns=['Symbol', 'CandleType', 'h1CandleType1', 'h2CandleType1'],
prior_weight=3, suffix='')), prior=None),
'enc__time__cyclicity': Categorical(categories=(CyclicalFeatures(drop_original=True),
CycleTransformer(), RepeatingBasisFunction(n_periods=96)), prior=None),
'dim__fs_wrapper': Categorical(categories=('passthrough',
SelectFromModel(estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_'),
RFECV(cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877, n_jobs=1,
num_leaves=59, random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_', min_features_to_select=10, n_jobs=1,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1), step=3),
GeneticSelectionCV(caching=True,
cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
mutation_proba=0.1, n_gen_no_change=3, n_generations=20, n_population=50,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1))), prior=None),
'clf__base_estimator__eval_metric': Categorical(categories=('logloss', 'aucpr'), prior=None),
'clf__base_estimator__max_depth': Integer(low=2, high=8, prior='uniform', transform='identity'),
'clf__base_estimator__min_child_weight': Real(low=1e-05, high=1000, prior='log-uniform', transform='identity'),
'clf__base_estimator__colsample_bytree': Real(low=0.1, high=1.0, prior='uniform', transform='identity'),
'clf__base_estimator__subsample': Real(low=0.1, high=0.9999999999999999, prior='uniform', transform='identity'),
'clf__base_estimator__learning_rate': Real(low=1e-05, high=1, prior='log-uniform', transform='identity'),
'clf__base_estimator__gamma': Real(low=1e-06, high=1000, prior='log-uniform', transform='identity')}
To Reproduce
Steps to reproduce the behavior:
<<< Please let me know if you would like more information to reproduce the error >>>
Expected behavior
On occasions when it ran successfully, I got the following results for best_params_ as expected:
AssertionError Traceback (most recent call last)
Input In [130], in <cell line: 7>()
2 start = time()
3 # sv_results_ray = cross_val_clone(ray_pipe, X, y, cv_val[VAL], result_metrics, #scoring=score_metrics,
4 # return_estimator=True, return_train_score=True,
5 # optimise_threshold=True, granularity=THRESHOLD_GRAN
6 # )
----> 7 sv_results_ray = cross_val_thresh(ray_pipe, X, y, cv_val[VAL], result_metrics, #scoring=score_metrics,
8 return_estimator=True, return_train_score=True,
9 thresh_split=SPLIT,
10 )
11 end = time()
Input In [46], in cross_val_thresh(estimator, X, y, cv, result_metrics, return_estimator, return_train_score, thresh_split, *args, **kwargs)
25 time_df.loc[i, 'split'] = i
27 start = time()
---> 28 est_i.fit(X_train, y_train, **kwargs) # **kwargs used to push callbacks to gen_pipe
29 end = time()
30 time_df.loc[i, 'fit_time'] = end - start
File C:\Anaconda3\envs\skl_py38\lib\site-packages\tune_sklearn\tune_basesearch.py:622, in TuneBaseSearchCV.fit(self, X, y, groups, tune_params, **fit_params)
597 def fit(self, X, y=None, groups=None, tune_params=None, **fit_params):
598 """Run fit with all sets of parameters.
599
600 tune.run is used to perform the fit procedure.
(...)
620
621 """
--> 622 return self._fit(X, y, groups, tune_params, **fit_params)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\tune_sklearn\tune_basesearch.py:589, in TuneBaseSearchCV._fit(self, X, y, groups, tune_params, **fit_params)
587 refit_start_time = time.time()
588 if y is not None:
--> 589 self.best_estimator.fit(X, y, **fit_params)
590 else:
591 self.best_estimator.fit(X, **fit_params)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:378, in Pipeline.fit(self, X, y, **fit_params)
352 """Fit the model.
353
354 Fit all the transformers one after the other and transform the
(...)
375 Pipeline with fitted steps.
376 """
377 fit_params_steps = self._check_fit_params(**fit_params)
--> 378 Xt = self._fit(X, y, **fit_params_steps)
379 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)):
380 if self._final_estimator != "passthrough":
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:336, in Pipeline._fit(self, X, y, **fit_params_steps)
334 cloned_transformer = clone(transformer)
335 # Fit or load from cache the current transformer
--> 336 X, fitted_transformer = fit_transform_one_cached(
337 cloned_transformer,
338 X,
339 y,
340 None,
341 message_clsname="Pipeline",
342 message=self._log_message(step_idx),
343 **fit_params_steps[name],
344 )
345 # Replace the transformer of the step with the fitted
346 # transformer. This is necessary when loading the transformer
347 # from the cache.
348 self.steps[step_idx] = (name, fitted_transformer)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:594, in MemorizedFunc.call(self, *args, **kwargs)
593 def call(self, *args, **kwargs):
--> 594 return self._cached_call(args, kwargs)[0]
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:537, in MemorizedFunc._cached_call(self, args, kwargs, shelving)
534 must_call = True
536 if must_call:
--> 537 out, metadata = self.call(*args, **kwargs)
538 if self.mmap_mode is not None:
539 # Memmap the output at the first call to be consistent with
540 # later calls
541 if self._verbose:
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\memory.py:779, in MemorizedFunc.call(self, *args, **kwargs)
777 if self._verbose > 0:
778 print(format_call(self.func, args, kwargs))
--> 779 output = self.func(*args, **kwargs)
780 self.store_backend.dump_item(
781 [func_id, args_id], output, verbose=self._verbose)
783 duration = time.time() - start_time
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:1154, in FeatureUnion.fit_transform(self, X, y, **fit_params)
1133 def fit_transform(self, X, y=None, **fit_params):
1134 """Fit all transformers, transform the data and concatenate results.
1135
1136 Parameters
(...)
1152 sum of n_components (output dimension) over transformers.
1153 """
-> 1154 results = self._parallel_func(X, y, fit_params, _fit_transform_one)
1155 if not results:
1156 # All transformers are None
1157 return np.zeros((X.shape[0], 0))
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:1176, in FeatureUnion._parallel_func(self, X, y, fit_params, func)
1173 self._validate_transformer_weights()
1174 transformers = list(self._iter())
-> 1176 return Parallel(n_jobs=self.n_jobs)(
1177 delayed(func)(
1178 transformer,
1179 X,
1180 y,
1181 weight,
1182 message_clsname="FeatureUnion",
1183 message=self._log_message(name, idx, len(transformers)),
1184 **fit_params,
1185 )
1186 for idx, (name, transformer, weight) in enumerate(transformers, 1)
1187 )
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:1043, in Parallel.call(self, iterable)
1034 try:
1035 # Only set self._iterating to True if at least a batch
1036 # was dispatched. In particular this covers the edge
(...)
1040 # was very quick and its callback already dispatched all the
1041 # remaining jobs.
1042 self._iterating = False
-> 1043 if self.dispatch_one_batch(iterator):
1044 self._iterating = self._original_iterator is not None
1046 while self.dispatch_one_batch(iterator):
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:861, in Parallel.dispatch_one_batch(self, iterator)
859 return False
860 else:
--> 861 self._dispatch(tasks)
862 return True
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:779, in Parallel._dispatch(self, batch)
777 with self._lock:
778 job_idx = len(self._jobs)
--> 779 job = self._backend.apply_async(batch, callback=cb)
780 # A job can complete so quickly than its callback is
781 # called before we get here, causing self._jobs to
782 # grow. To ensure correct results ordering, .insert is
783 # used (rather than .append) in the following line
784 self._jobs.insert(job_idx, job)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib_parallel_backends.py:208, in SequentialBackend.apply_async(self, func, callback)
206 def apply_async(self, func, callback=None):
207 """Schedule a func to be run"""
--> 208 result = ImmediateResult(func)
209 if callback:
210 callback(result)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib_parallel_backends.py:572, in ImmediateResult.init(self, batch)
569 def init(self, batch):
570 # Don't delay the application, to avoid keeping the input
571 # arguments in memory
--> 572 self.results = batch()
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:262, in BatchedCalls.call(self)
258 def call(self):
259 # Set the default nested backend to self._backend but do not set the
260 # change the default number of processes to -1
261 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 262 return [func(*args, **kwargs)
263 for func, args, kwargs in self.items]
File C:\Anaconda3\envs\skl_py38\lib\site-packages\joblib\parallel.py:262, in (.0)
258 def call(self):
259 # Set the default nested backend to self._backend but do not set the
260 # change the default number of processes to -1
261 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 262 return [func(*args, **kwargs)
263 for func, args, kwargs in self.items]
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\utils\fixes.py:117, in _FuncWrapper.call(self, *args, **kwargs)
115 def call(self, *args, **kwargs):
116 with config_context(**self.config):
--> 117 return self.function(*args, **kwargs)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\sklearn\base.py:870, in TransformerMixin.fit_transform(self, X, y, **fit_params)
867 return self.fit(X, **fit_params).transform(X)
868 else:
869 # fit method of arity 2 (supervised transformation)
--> 870 return self.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:279, in GeneticSelectionCV.fit(self, X, y, groups)
262 def fit(self, X, y, groups=None):
263 """Fit the GeneticSelectionCV model and then the underlying estimator on the selected
264 features.
265
(...)
277 instance (e.g., GroupKFold).
278 """
--> 279 return self._fit(X, y, groups)
File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:343, in GeneticSelectionCV._fit(self, X, y, groups)
340 print("Selecting features with genetic algorithm.")
342 with np.printoptions(precision=6, suppress=True, sign=" "):
--> 343 _, log = _eaFunction(pop, toolbox, cxpb=self.crossover_proba,
344 mutpb=self.mutation_proba, ngen=self.n_generations,
345 ngen_no_change=self.n_gen_no_change,
346 stats=stats, halloffame=hof, verbose=self.verbose)
347 if self.n_jobs != 1:
348 pool.close()
File C:\Anaconda3\envs\skl_py38\lib\site-packages\genetic_selection\gscv.py:50, in _eaFunction(population, toolbox, cxpb, mutpb, ngen, ngen_no_change, stats, halloffame, verbose)
48 fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
49 for ind, fit in zip(invalid_ind, fitnesses):
---> 50 ind.fitness.values = fit
52 if halloffame is None:
53 raise ValueError("The 'halloffame' parameter should not be None.")
File C:\Anaconda3\envs\skl_py38\lib\site-packages\deap\base.py:188, in Fitness.setValues(self, values)
187 def setValues(self, values):
--> 188 assert len(values) == len(self.weights), "Assigned values have not the same length than fitness weights"
189 try:
190 self.wvalues = tuple(map(mul, values, self.weights))
AssertionError: Assigned values have not the same length than fitness weights
However, when I exclude dim__fs_wrapper from the pipeline, the error does not occur at all. The purpose of this transformer is to select a feature selection method from amongst 'passthrough' and estimators wrapped in SelectFromModel, RFECV and GeneticSelectionCV and the error seems to originate when GeneticSelectionCV is used for feature selection.
Additional context
- Note that my approach involves packaging all transformers and classifier within the pipeline and running hyperparameter tuning upon all elements of the pipeline rather than on the classifier only. This allows me to select from many different transformers for the same purpose e.g. target encoding, dimension reduction etc., instead of limiting myself to just one.



