Overview
The Open Graph Benchmark (OGB) is a collection of benchmark datasets, data loaders, and evaluators for graph machine learning. Datasets cover a variety of graph machine learning tasks and real-world applications. The OGB data loaders are fully compatible with popular graph deep learning frameworks, including PyTorch Geometric and Deep Graph Library (DGL). They provide automatic dataset downloading, standardized dataset splits, and unified performance evaluation.
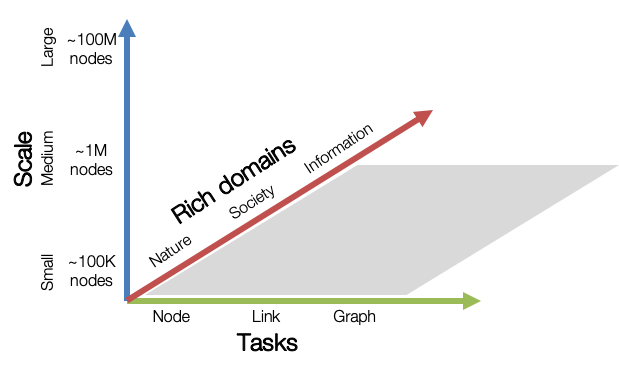
OGB aims to provide graph datasets that cover important graph machine learning tasks, diverse dataset scale, and rich domains.
Graph ML Tasks: We cover three fundamental graph machine learning tasks: prediction at the level of nodes, links, and graphs.
Diverse scale: Small-scale graph datasets can be processed within a single GPU, while medium- and large-scale graphs might require multiple GPUs or clever sampling/partition techniques.
Rich domains: Graph datasets come from diverse domains ranging from scientific ones to social/information networks, and also include heterogeneous knowledge graphs.
OGB is an on-going effort, and we are planning to increase our coverage in the future.
Installation
You can install OGB using Python's package manager pip. If you have previously installed ogb, please make sure you update the version to 1.3.2. The release note is available here.
Requirements
- Python>=3.6
- PyTorch>=1.6
- DGL>=0.5.0 or torch-geometric>=1.6.0
- Numpy>=1.16.0
- pandas>=0.24.0
- urllib3>=1.24.0
- scikit-learn>=0.20.0
- outdated>=0.2.0
Pip install
The recommended way to install OGB is using Python's package manager pip:
pip install ogb
python -c "import ogb; print(ogb.__version__)"
# This should print "1.3.2". Otherwise, please update the version by
pip install -U ogb
From source
You can also install OGB from source. This is recommended if you want to contribute to OGB.
git clone https://github.com/snap-stanford/ogb
cd ogb
pip install -e .
Package Usage
We highlight two key features of OGB, namely, (1) easy-to-use data loaders, and (2) standardized evaluators.
(1) Data loaders
We prepare easy-to-use PyTorch Geometric and DGL data loaders. We handle dataset downloading as well as standardized dataset splitting. Below, on PyTorch Geometric, we see that a few lines of code is sufficient to prepare and split the dataset! Needless to say, you can enjoy the same convenience for DGL!
from ogb.graphproppred import PygGraphPropPredDataset
from torch_geometric.data import DataLoader
# Download and process data at './dataset/ogbg_molhiv/'
dataset = PygGraphPropPredDataset(name = 'ogbg-molhiv')
split_idx = dataset.get_idx_split()
train_loader = DataLoader(dataset[split_idx['train']], batch_size=32, shuffle=True)
valid_loader = DataLoader(dataset[split_idx['valid']], batch_size=32, shuffle=False)
test_loader = DataLoader(dataset[split_idx['test']], batch_size=32, shuffle=False)
(2) Evaluators
We also prepare standardized evaluators for easy evaluation and comparison of different methods. The evaluator takes input_dict (a dictionary whose format is specified in evaluator.expected_input_format) as input, and returns a dictionary storing the performance metric appropriate for the given dataset. The standardized evaluation protocol allows researchers to reliably compare their methods.
from ogb.graphproppred import Evaluator
evaluator = Evaluator(name = 'ogbg-molhiv')
# You can learn the input and output format specification of the evaluator as follows.
# print(evaluator.expected_input_format)
# print(evaluator.expected_output_format)
input_dict = {'y_true': y_true, 'y_pred': y_pred}
result_dict = evaluator.eval(input_dict) # E.g., {'rocauc': 0.7321}
Citing OGB / OGB-LSC
If you use OGB or OGB-LSC datasets in your work, please cite our papers (Bibtex below).
@article{hu2020ogb,
title={Open Graph Benchmark: Datasets for Machine Learning on Graphs},
author={Hu, Weihua and Fey, Matthias and Zitnik, Marinka and Dong, Yuxiao and Ren, Hongyu and Liu, Bowen and Catasta, Michele and Leskovec, Jure},
journal={arXiv preprint arXiv:2005.00687},
year={2020}
}
@article{hu2021ogblsc,
title={OGB-LSC: A Large-Scale Challenge for Machine Learning on Graphs},
author={Hu, Weihua and Fey, Matthias and Ren, Hongyu and Nakata, Maho and Dong, Yuxiao and Leskovec, Jure},
journal={arXiv preprint arXiv:2103.09430},
year={2021}
}