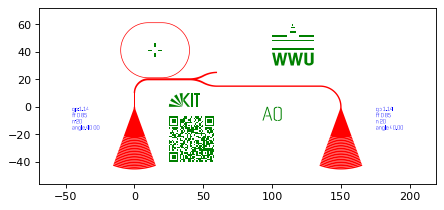
GDSHelpers
GDSHelpers in an open-source package for automatized pattern generation for nano-structuring. It allows exporting the pattern in the GDSII-format and OASIS-format, which are currently mainly used for describing 2D-masks. Currently, the focus is mainly on photonic and superconducting circuitry. The library consists of growing list of parts, which can be composed into larger circuits.
So far, the following parts are implemented:
- A waveguide part, allowing easy chaining of bends and straight waveguides.
- Includes parameterized paths and Bézier curves.
- Automatic smooth connection to a target point/port
- The size of the waveguide can be tapered (linear or by a user defined function), which can e.g. be used for optical edge coupling or electronic contact pads
- Allows to design slot-waveguides and coplanar waveguides (with arbitrary number of rails)
- Different types of splitters:
- Y-splitter
- MMI-splitters
- Directional splitter
- Couplers
- Grating couplers (allowing apodized gratings)
- Tapers for hybrid 3D-integration
- Ring and racetrack resonators
- Mach-Zehnder interferometers
- Strip to slot mode converter
- Spirals
- Superconducting nanowire single photon detectors (SNSPDs)
- Superconducting nanoscale Transistors (NTRONs)
- Different types of markers
- QRcodes
- A possibility to include images
- Text-elements for labeling the structures
- GDSII-import
Besides this it also allows to perform conveniently operations on the design, like:
- Convert the pattern for usage of positive resist
- Create holes around the circuitry, which is e.g. necessary for under-etching
- Shapely-operations can also be applied on the generated structures, e.g. shrinking or inflating of the geometry
- Surrounding the structures with holes in a rectangular lattice and filling waveguides with holes in a honeycomb lattice for controlling vortex dynamics
The structures are organized in cells, which allow:
- Adding structures on multiple layers
- Adding cells into other cells, the cells can be added with an offset with respect to the parent cell and can be rotated
- Storing additional information, which can be used for saving design parameters
- Automatized generation of region layers
- Parallelized export
Additionally the structures can conveniently be simulated by:
- Using the meep integration (FDTD)
Finally, there are also different formats in which the pattern can be exported:
- The GDSII-format, which is quite often used for (electron beam/...)-lithography
- The OASIS-format, which one of the successors of the GDSII-format
- To an 2D-image
- To stl-objects which are useful e.g. for 3D-renderings
- Directly to a blender-file or an rendered 3D-image
Citing GDSHelpers
We would appreciate if you cite the following paper in your publications for which you used GDSHelpers:
Helge Gehring, Matthias Blaicher, Wladick Hartmann, and Wolfram H. P. Pernice, "Python based open source design framework for integrated nanophotonic and superconducting circuitry with 2D-3D-hybrid integration" OSA Continuum 2, 3091-3101 (2019)
Documentation
You can find the documentation on readthedocs. If you have problems using GDSHelpers don't hesitate to contact us using Discussions or send me a mail.
Installation
The GDSHelpers can be installed via pip using (more details in the installation documentation)
pip install gdshelpers