PyPi
Conda

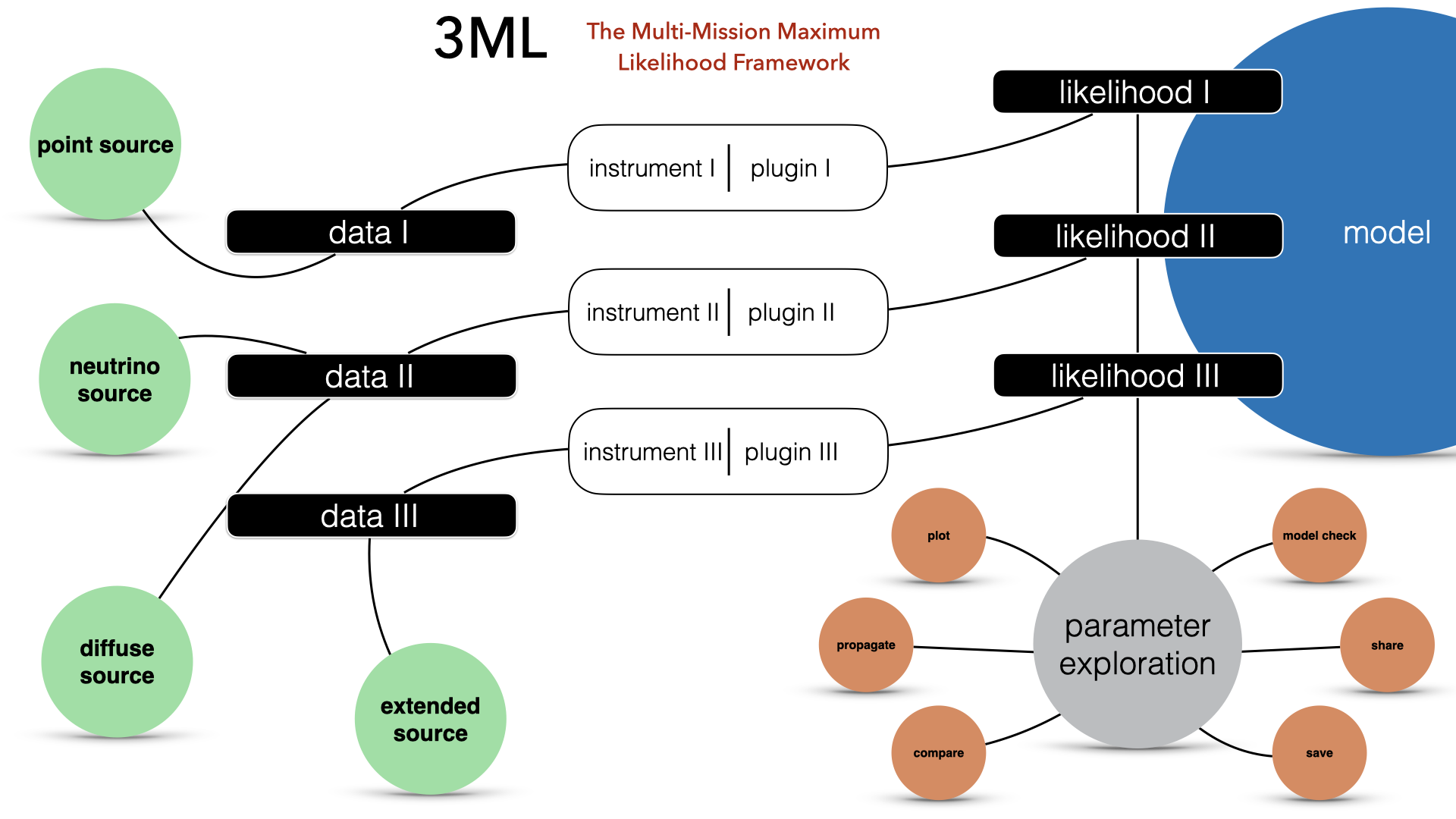
The Multi-Mission Maximum Likelihood framework (3ML)
A framework for multi-wavelength/multi-messenger analysis for astronomy/astrophysics.
Astrophysical sources are observed by different instruments at different wavelengths with an unprecedented quality. Putting all these data together to form a coherent view, however, is a very difficult task. Indeed, each instrument and data type has its own ad-hoc software and handling procedure, which present steep learning curves and do not talk to each other.
The Multi-Mission Maximum Likelihood framework (3ML) provides a common high-level interface and model definition, which allows for an easy, coherent and intuitive modeling of sources using all the available data, no matter their origin. At the same time, thanks to its architecture based on plug-ins, 3ML uses under the hood the official software of each instrument, the only one certified and maintained by the collaboration which built the instrument itself. This guarantees that 3ML is always using the best possible methodology to deal with the data of each instrument.
Though Maximum Likelihood is in the name for historical reasons, 3ML is an interface to several Bayesian inference algorithms such as MCMC and nested sampling as well as likelihood optimization algorithms. Each approach to analysis can be seamlessly switched between allowing users to try different approaches quickly and without having to rewrite their model or data interfaces.
Like your XPSEC models? You can use them in 3ML as well as our growing selection of 1-,2- and 3-D models from our fast and customizable modeling language astromodels.
Installation
Installing with pip or conda is easy
pip install astromodels threeml
conda install astromodels threeml -c threeml conda-forge
Please refer to the Installation instructions for more details and trouble-shooting.
Press
Who is using 3ML?
Here is a highlight list of teams and their publications using 3ML.
A full list of publications using 3ML is here.
Citing
If you find this package useful in you analysis, or the code in your own custom data tools, please cite:
Acknowledgements
3ML makes use of the Spanish Virtual Observatory's Filter Profile service (http://svo2.cab.inta-csic.es/svo/theory/fps3/index.php?mode=browse&gname=NIRT).
If you use these profiles in your research, please consider citing them by using the following suggested sentence in your paper:
"This research has made use of the SVO Filter Profile Service (http://svo2.cab.inta-csic.es/theory/fps/) supported from the Spanish MINECO through grant AyA2014-55216"
and citing the following publications:
The SVO Filter Profile Service. Rodrigo, C., Solano, E., Bayo, A. http://ivoa.net/documents/Notes/SVOFPS/index.html The Filter Profile Service Access Protocol. Rodrigo, C., Solano, E. http://ivoa.net/documents/Notes/SVOFPSDAL/index.html